Psychiatric Genetics Beyond Heritability: Q&A with Michel Nivard
We look for genes as a means to an end—biology, epidemiology, and etiology of complex human outcomes.
Michel Nivard, PhD, is a professor of genetic epidemiology at the MRC Integrative Epidemiology Unit in Bristol, UK. He was trained as a psychologist at the University of Amsterdam and has a PhD in psychiatric genetics from the Vrije Universiteit Amsterdam. He writes online on Michel’s Substack.
Awais Aftab, MD, is a clinical assistant professor of psychiatry at Case Western Reserve University. He is interested in conceptual and philosophical issues in psychiatry. His first book, Conversations in Critical Psychiatry (OUP, 2024), is an edited collection of interviews.
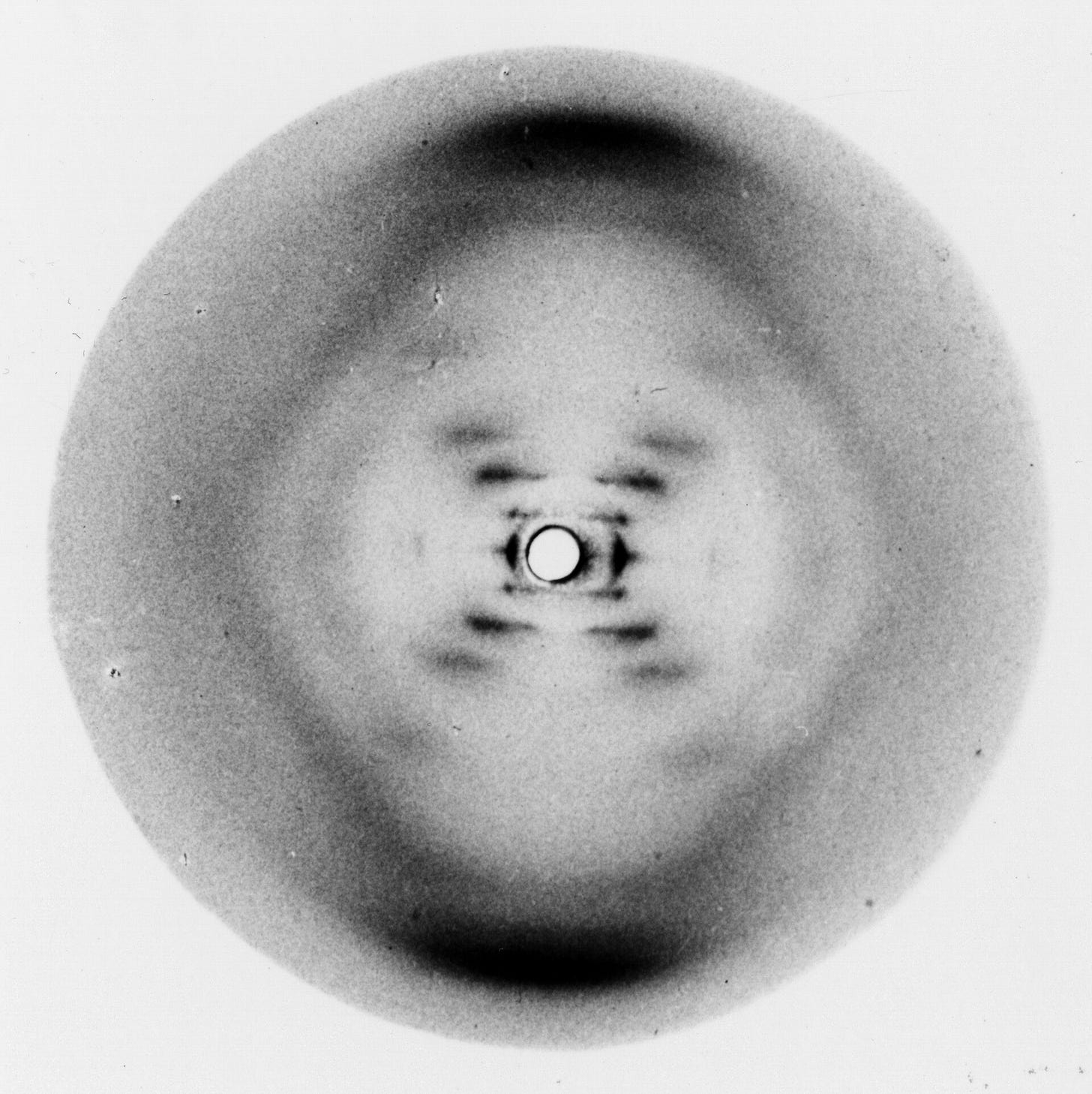
Aftab: Psychiatric genetics has been a recurring topic on Psychiatry at the Margins, and I keep returning to it because it feels like an itch I can’t quite scratch. A lot of it, I suspect, has to do with the fact that from some vantage points, genetics appears to have an outsized importance to psychiatry (high twin heritabilities, for example), but from other vantage points, it barely seems to matter (low SNP heritabilities, low predictive value of polygenic risk scores [PRS] for most conditions). Among contemporary psychopathology researchers, I sense a quiet resignation, an acceptance that genetics may be important in some abstract sense, but in concrete terms, in terms of identifiable genes and direct genetic effects, it isn’t going to provide us with adequate etiological explanations or have a transformative impact on clinical care. How do you read the current mood in the psychopathology and clinical psychology communities?
Nivard: So perhaps instead of diving into the numbers, high heritability, low SNP heritability, etcetera, it’s good to consider why we’d want to know, or whether we should care. See, the Human Genome Project was envisioned as a tool to systematically link biology, across all levels, and perhaps even mental processes like cognitions, to complex outcomes like psychiatric diagnoses. We don’t study the genome for its own sake, especially since when the Human Genome Project played out, intervening on the genome must have sounded like sci-fi. Rather, the thinking was, and is, that if we find genes that relate to being diagnosed with, say, schizophrenia, we can trace those genes’ effects deep into biology. Those genes might be expressed in certain tissues (neurons), in specific developmental phases (for example, prenatal), and are involved in specific neuronal functions or systems.
Now, the genome doesn’t owe us a specific answer when we interrogate it to study the etiology of something as complex as schizophrenia or bipolar disorder. There are immune theories of schizophrenia, so it could have been that all those genes would be expressed in immune cells, or in microglia, cells that scavenge the brain for plaques or damaged neurons. Whatever the etiology of an outcome, finding the genes involved will let you trace that etiology.
It’s also the case that the general outlines of the answer were becoming clear early on. The first solid estimate of the SNP heritability of schizophrenia came in 2009 and was based on then-revolutionary polygenic scores. That work estimated the SNP heritability at about 30% and, more intriguingly, flagged that about 5–10% of the whole genome had some relation to schizophrenia. Everyone treated PRS as a potential tool for clinical prediction after that paper, but the most interesting finding was that the great majority of the total genetic effects on schizophrenia were distributed across a staggering number of variants, each of which has a small effect. This means that for most individual patients, genetic influences on their mental health are the sum of the effects of genetic variants with medium or small effects distributed widely across the genome. Meaning, for those individuals there is no unitary cause, which I personally believe is a great etiological insight. We have further been slowly mapping that genetic signal to neurons and, to some extent, to certain developmental phases and functions and systems within neurons, but mostly we have found that a great many systems in the brain, specifically those related to neurons, are implicated in being diagnosed with schizophrenia. Again, those efforts haven’t pointed at a very specific process, developmental phase, or region in the brain, or in the neuron, as the root cause of schizophrenia. The genetic variants that relate to schizophrenia are many, which likely means there are many diffuse contributory causes. I don’t think that finding is a failure of psychiatric genetics
Now those common variants do not explain the total heritability. There are also a few dozen genes in which rare deleterious genetic variants, mutations that directly corrupt a gene we know to be intolerant to being corrupted, are known to considerably change your risk for severe mental health issues. A small percentage of people diagnosed with schizophrenia will carry one of these rare, very deleterious variants. For these individuals, it’s plausible that without the variant, they wouldn’t have had mental suffering to the point where they’d end up in care.
There are obviously also environmental causes. I wrote a book chapter on the environmental causes of psychopathology, and there are plenty. For schizophrenia, migration in critical developmental ages might be one, severe trauma another. But a specific type of cause (can be genetic or environmental) is often overlooked: near-ubiquitous causes. The head of my department, George Davey-Smith, frequently encourages me to consider their role; he wrote on their potential role in cancer epidemiology. A relevant example to consider is Multiple Sclerosis: to develop it, it appears you must have been exposed to Epstein-Barr Virus. But nearly everyone has been exposed to EBV. EBV is a necessary (you must have had it) but insufficient (not everyone who had EBV gets MS) cause of MS. If you GWAS MS, as people did, you get a heritability, you get all kinds of GWAS hits, and while a portion of those genome wide hits might be related to susceptibility to EBV that won’t be immediately obvious and might have escaped us if we hadn’t known EBV existed.
In a way, we have had similar situations in psychiatry historically. I imagine a great portion of people with psychosis in the 19th century in fact had neurosyphilis, which can present with psychosis, but we don’t currently consider those people to have schizophrenia; we treat the syphilis. There is a non-zero chance that there are other ubiquitous exposures that are the root cause of disease for a portion of patients with psychosis. If that is the case, a portion of the heritability of schizophrenia is then related to individual differences in susceptibility to that ubiquitous exposure, and GWAS might be a way to try and find such ubiquitous exposures.
Aftab: A common critical narrative about psychiatric genetics goes something like this: researchers claimed that mental disorders were genetic diseases caused by abnormal genes, spent years searching for these faulty genes, failed to find them, and are now reluctant to admit that the project has not delivered on its original promises. Within the field, of course, there’s widespread acknowledgment that psychiatric disorders are highly polygenic, involving thousands of variants with small, nonspecific, and often transdiagnostic effects. Still, the sweeping questions about “genes for mental illness” that dominated public and professional debate in the late 20th century seem to have been quietly set aside. Today, psychiatric geneticists appear more focused on what can be practically done with genetic data: improving prediction through polygenic scores, potentially informing personalized treatment, and identifying new drug targets. All of this presumes that genes matter in non-trivial ways, even if they are not central to causation in the classical sense of genetic abnormalities.
As I was working on these questions, Eric Turkheimer commented on the new GWAS of personality paper and noted that the goal of disentangling genetic and environmental influences has been largely abandoned. He also suggested that the field has stopped trying to identify the “genes for” anything in particular, instead being content with reporting significant SNP correlations and characteristics.
How would you characterize the shift in the overarching goals of psychiatric genetics from the 1990s to the 2020s? And does the field need to publicly reckon with any perceived failures or unrealized expectations?
Nivard: First of all, I was 7 in 1990, so I feel no need to reckon with any perceived failures of unrealized expectations of the field writ large. I have my hands full with my own very real self-doubts as a scientist. I just don’t know if there is, or ever was, such a thing as “the field.” A key critique of GWAS in like 2007-2009 was that early results implied that massive portions of the genome would be involved in complex traits; the pioneers of psychiatric genetics themselves empirically confirmed that by 2009. I think this led some of those pioneers to pivot to rare variants and others to try and develop tools to deal with the reality of high polygenicity. Looking back, it seems to me the die had been cast on the next decade of GWAS by 2009. Now that’s me looking back; you can pick an entirely different narrative! Hindsight and our current perspectives distort our view of the past, certainly mine. One thing I deeply appreciate about genetics is that people do in fact cumulatively build knowledge. So if ideas are quietly set aside, that’s a consequence of the bulk of the field accepting empirical results and proceeding on the basis of those findings.
Nivard: One thing I deeply appreciate about genetics is that people do in fact cumulatively build knowledge. So if ideas are quietly set aside, that’s a consequence of the bulk of the field accepting empirical results and proceeding on the basis of those findings.
That being said, I think the entire frame “quantify genes or the environment” is the wrong frame. We don’t look for genes as an end; rather, we look for genes as a means to an end. Those ends are: biology, epidemiology, and a complete etiology of complex human outcomes. If we find a gene that affects LDL, or smoking, or schizophrenia diagnoses, we can trace their effects in cells, in organs, or systems, or on other outcomes related to our outcome of interest to begin building up a complete picture of the causal web that results in a diagnosis of schizophrenia.
The top hit of a lung cancer GWAS is in the nicotinic receptor gene, because smoking causes lung cancer. Had we not known smoking causes lung cancer, we could have used the GWAS to find out. A GWAS of lung cancer in Europe in 1492, before Columbus departed, would not have had a lead hit in the nicotinic receptor gene because nicotine is a New World plant, and so people didn’t smoke tobacco in Europe. A GWAS of lung cancer in 3120 hopefully won’t have a hit in the nicotinic receptor gene because policy will have resulted in no or very few smokers.
A GWAS isn’t always a reflection of biology; it’s whatever massive, unwieldy causal process, be it sociological, policy-related, psychological, or biological, results in some people ending up in the case group and others in the control group. A GWAS of a diagnosis will always reflect the etiology of being in that diagnostic category, conditional on the sample, society, and contemporary environmental situation. A GWAS doesn’t owe us a specific, easily digestible etiological story; trying to insist on one is the wrong way to approach genetic epidemiology.
A great early illustration of how we can leverage genetics to highlight, and perhaps mitigate, the messiness of observational epidemiology was this paper by Joanna Martin, who showed that polygenic risk scores for schizophrenia are associated with mothers and their children dropping out of longitudinal cohort studies. Knowing who isn’t in your study is the first tiny step to mitigating the fact that some people aren’t in it.
The reason that a GWAS, which is generally messy and observational, can still be very useful is because your genetics are set at birth. So if a gene correlates to an outcome, we can rule out that the outcome might have caused the gene. With a little more work (controlling for sources of gene–environment correlation), we can establish with some certainty that a gene causes an average change in the outcome. This then lets us do interesting natural experiments.
Let me give you one example that is relevant to psychiatry. Vitamin D levels have long been linked to depression, but you’ve got to wonder whether that is the cause of depression, the consequence, or merely correlated, right? People did careful longitudinal epidemiology which didn’t rule out an effect, and eventually did very large trials, in which they found no effect of vitamin D supplementation in the general population on depression over a 5-year period.
In retrospect, genetic epidemiology could have avoided those very expensive trials. There are a few variants with large effects on circulating vitamin. These effects are direct, and their biology is well understood. If vitamin D levels are causal for depression, then the variants that cause vitamin D levels to go up or down must cause depression risk to go down. They do not, and so we can rule out a causal role for vitamin D in the etiology of depression.
Now I want to also clearly articulate some risks that arise from this kind of genetic epidemiology. The ability to take any two GWAS and automatically run the statistical machinery to test whether a causal effect is plausible, combined with perverse publishing incentives for academics and MDs, led to a deluge of thoughtless papers. People just take a random GWAS of a protein or metabolite, a GWAS of a disease or trait B, run a little test, then retroactively cobble together an introduction, write a paper, and submit. Add in the bias towards publishing significant findings and you get a very potent noise machine.
Doing good science, getting value from our massive collective investments in (genetic) epidemiology, is hard and requires a level of seriousness. It cannot just rely on automated and scripted processes for finding truth.
That being said, the genetic epidemiology in which we move beyond wondering about “genes or environment?” and use genes to study (environmental or biological) causes and consequences of mental health outcomes, whatever they may be, is the real deal.
Of course, genetic epidemiology has its blind spots beyond careless application. A key blind spot is the human element to it all. I hope I made abundantly clear that we could find all kinds of etiological clues in GWAS, not just biological, but the choice to interrogate a GWAS using tools to uncover biology, or to try and parse out socially or psychologically mediated etiological processes, is a human choice the researcher makes. There are no universally agreed-upon tests we can subject a GWAS to in order to determine whether seeking out answers in biology, psychology, or sociology is going to be most fruitful.
You’ll notice I haven’t spoken about risk prediction at all, but here similar dynamics are at play. There is useful prediction, and there is senseless prediction. Psychiatric services everywhere are strained as is; what would prediction help solve?
Say we have a predictor that can substitute the general risk for bipolar disorder (which is ~1%) for a range between 0.05% and 20%, then what? Experience teaches us that the risk would be distributed near normally, with most people bunched near 1%. So for those people, nothing changes. Some people might be at the low end, say <0.1%, more than 10 times less than the average risk. Those people still wouldn’t live their lives differently because few of us worry about low-risk future events. But perhaps people with relatives with bipolar disorder would be relieved. People with higher risk, say 10–20%, could be intervened on, but how? We don’t really have long-term preventative care for bipolar disorder. We could raise awareness of early signs, but do we know for sure that wouldn’t just cause tremendous stress and worry?
Geneticists cannot design and implement the risk prediction programs you’d need. Psychiatrists, in concert with epidemiologists and geneticists, might. I am a huge fan of recent work by Dutch psychiatrist Wouter Peyrot that does just that. As a psychiatrist, he first homed in on a very real situation psychiatrists in acute care contend with: you come across a new patient, young, no prior history, with severe mood-related symptoms, is this as bad as it gets? Will it become MDD, or might they eventually develop manic or psychosis-like symptoms? Do you weigh those risks in your treatment decisions?
See, in this selected group of patients, the prior risk of bipolar disorder isn’t 1%, but depending on the care system and type of clinic, maybe 10, 15, or even 25%. In this scenario, polygenic scores might actually be very useful to assess risk, because a high score in this group might move the 15% prior risk to, say, 60% risk of bipolar. At that point, your treatment choices might change, or you might structure this individual and their environment to be aware of early signs of mania and give them instructions on how to act.
For risk prediction to be fruitful in this group would require policy and implementation studies, clinician training, constant routine outcome monitoring, efficient and cheap bioinformatic pipelines, and it would require us to improve PRS for individuals that aren’t of broadly European ancestry (because most of our GWAS data is biased towards that group). All those goals are worthwhile but obviously should be considered relative to other care innovations.
Should we spend considerable resources on this? Or on more tightly integrating psychiatric care and municipal housing services? I don’t know, but I know both are potentially fruitful efforts that would require sustained effort and funding, and opting for one might mean there is no budget for another.
Scientists and clinician-scientists are good at dreaming up prediction models. We aren’t as good at effectively implementing them, even as deciding whether we should implement them is an incredibly hard question that requires deep policy knowledge of the whole set of potentially conflicting policy goals. This isn’t a failure of genetic prediction (nor a success!) in specific, but a failure of academia writ large to translate probabilistic information into medically relevant and actionable information.
You might pick up on a common theme here: I don’t view psychiatric genetics as “obviously worth it” or “oversold” based on the empirical literature. There is amazing work out there, and there is noise out there. The amazing work needs brilliant biological translation and eventually clinical implementation, and I am optimistic some of that will happen over time.
Aftab: What’s your perspective on the “missing heritability” problem in psychiatric genetics? Do you think it remains a central issue, or has the field moved past it in meaningful ways?
Nivard: Heritability estimates based on family and twin data were on the high end. Using molecular genetic data has allowed us to produce better, almost always lower, estimates of heritability. It’s not fruitful to directly compare those estimates, though; they’re different in the sense that the different models make different assumptions. In a twin model we essentially start our inquiry stating: “Assuming genes and the environment aren’t correlated, nor interact with each other; assuming people absolutely randomly choose their partners with no preference for anything heritable whatsoever; assuming all people in our diagnosed group are affected by the same disease (there is no diagnostic confusion); and assuming twins are in other ways interchangeable with the rest of the population.” For each newer model, there is a similar sort of preamble, a preamble most in the field are familiar with, but we don’t always spell out. All science requires assumptions. Assumptions are fine, but if two models have different assumptions, and those assumptions aren’t met perfectly, they’ll give you different results.
While in the day-to-day work of psychiatric geneticists the exact magnitude of heritability and the nuances of estimating it given a set of assumptions matter very little, other than to people like myself, who might want to study those assumptions and the processes they imply, heritability seems to matter greatly in non-scientific or popular scientific discourse.
My formative academic years coincided with a lot of genetically flavored scientific racism (I say “flavored” because most of these people really didn’t do science so much as advocacy for their views, dressed up with numbers). People wrote books based on the premise that IQ and education were 90% and 50% heritable, and that those numbers had important implications for society. In fact, they wanted to use those numbers, which had nothing to do with differences between racial groups, to justify or entertain genetic theories of racial differences in cognition, and for society to respond to those.
A decade later when the first GWAS of behavioral outcomes arrived, a lot of people were, understandably, very anxious about researchers doing GWAS of outcomes like education. It turned out that doing the science earnestly, and without a preconceived racial narrative, resulted in the field being able to show that education is not that heritable. Those studies also established that unmeasured gene-environment correlation, where parental genes correlate with child outcomes, biased earlier heritability estimates based on GWAS significantly. I am of the view that it was worth doing those GWAS earnestly, because they revealed a complex, nuanced process by which social stratification and partner choice shape educational outcomes and, in turn, affect our GWAS results.
However, we can’t pat ourselves on the back and let our guard down when it comes to misuse of our sciences. Genetics and eugenics are intertwined not by accident but because some of the leading 20th-century geneticists were eugenicists. The term “heritability” has taken on a life of its own, and because of it, we must choose our words carefully and realize that there is great appetite out there among ethno-nationalists to take genetics out of context and lean into old, deeply ingrained retrograde beliefs. I was a huge fan of US-funded efforts to improve high-school genetics curricula in a way that makes it harder to plant these ideas, and it worries me greatly that at least some of those efforts were defunded by the current US government.
As a scientist, I care very little about the absolute value of the statistical estimate of heritability for anything; it’s incredibly contextual and can’t really be decontextualized without collapsing in on itself. I gave various examples of rare, potentially protective, and actionable genes for Alzheimer’s, LDL cholesterol, schizophrenia, or nicotine dependence. These variants have enormous clinical potential while contributing tiny portions to the heritability of these outcomes. As someone following the political discourse closely, I have to care about what others think, or want to make you think, heritability means.
Nivard: As a scientist, I care very little about the absolute value of the statistical estimate of heritability for anything; it’s incredibly contextual and can’t really be decontextualized without collapsing in on itself.
Aftab: I interviewed Sasha Gusev last year about some foundational topics in behavioral genetics. Were there any points he raised in the discussion that you see differently or think deserve a different emphasis?
Nivard: So, in your interview Sasha highlighted that those rare variants with large effects, which he describes as “variants with small effects in the population,” might still point us towards general pathways on which to intervene that could improve outcomes for everyone. He highlighted the CHRNB2 gene; people who have rare deleterious variants in that gene smoke far less, and if the gene is knocked out in mice, they tend to get addicted to nicotine far less easily. Similarly, a gene (PCSK9) in which rare deleterious variants (deleterious for the gene, not the person) resulted in very low LDL cholesterol is being translated to very effective therapeutics. These examples of potential success derive from very rare protective variants whereby disabling the gene reduces LDL (or nicotine cravings). In these cases, disabling a gene, or somehow inactivating it through intervention, is expected to decrease LDL or craving for nicotine.
Now, it’s early days, and a GWAS is always observational. We have to figure out why these people smoke less; maybe the epithelium of their lungs is brittle and smoking feels unpleasant? If that’s the case, we wouldn’t want to try and develop drugs that target the same pathway as the gene, lest we risk making people's lungs brittle. However, protective genes are very potent leads for drug development.
In contrast, identifying and targeting genes where very rare deleterious variants increase the risk for schizophrenia would potentially only help those who carry deleterious variants in that gene. It’s very hard to identify protective variants among the 99% of individuals who will never develop psychosis, as what we’d really be looking for is a subset of individuals with a very low chance of ever developing schizophrenia. Most people never experience psychosis, not because they are in any way especially protected, but simply because it’s rare. In this majority, we’d be looking for a needle in a haystack, and we don’t even really understand what we’d be looking for. What would “very low risk for psychosis” present like?
The closest we have found to a protective variant is through our Copy Number Variant (CNV) studies. A CNV is a genetic variant where a long stretch of genome is duplicated (copied) or deleted, and people vary in the number of copies they carry. Some of the strongest risk variants for schizophrenia are CNVs. A deletion of a section of chromosome 22 (called chr22q11.2 deletion) strongly increases the risk of schizophrenia-like signs and symptoms; however, a duplication of the same region potentially decreases the risk of schizophrenia substantially. The authors that found the variant did worry that, because these kinds of large disruptions in the genome can cause syndrome-like presentations (seizures, developmental delay, etc.), perhaps there is a depletion of schizophrenia diagnoses because people are receiving care for other diagnoses, but early evidence suggests that’s not what is happening here.
There have been successful and less successful replications of this effect, so the jury is still out. I think we should give some serious consideration to testing whether we can psychometrically measure people who are exceedingly unlikely to develop psychosis and go back to qualitatively interview CNV duplication carriers and their families. We should use integrated national registers in Scandinavia to dig deep into the life course of these putative protective CNV carriers to establish whether they are truly protected or just simply shifted into other psychiatric diagnoses. At the same time, dig deep into the biology of genes affected by these rare protective CNVs. The chr22q11.2 CNV duplication raised the expression of over 30 genes that are on the duplicated segment of genome, among which is COMT, which plays a role in the breakdown of certain neurotransmitters. But obviously, it would be a mistake to presuppose it’s not one of the other genes. Look, even with leads like these, it will probably be very hard to resolve the causal mechanism, but if we succeed, then the odds of finding a way to intervene would go up.
It’s crucial to realize that there wouldn’t have been easy shortcuts had GWAS of schizophrenia turned out differently. If GWAS of schizophrenia had found a few genes with massive effects, following up on those would still have been very hard. By 1993, the HTT gene had been identified as the sole cause of Huntington’s disease, and the APOE gene was linked to Alzheimer’s. It’s fairly plausible that if everyone carried two copies of the rare low-risk APOE allele (ε2 ε2), 75% of Alzheimer’s patients would not have developed Alzheimer’s. Despite the very early identification of these genes with massive effects, there hasn’t been a therapeutic breakthrough for either disease yet. While therapeutics for high LDL cholesterol, based on intervening on a pathway influenced by a gene found through a protective rare variant, are on the market (and others are in development), the individual gene effects on LDL are far smaller than the effects of APOE on Alzheimer’s. Massive effect genes or high heritability aren’t necessary or sufficient conditions for therapeutic success. This is why, to me, the entire debate about high or low heritability is interesting but not relevant to the chances psychiatric genetics will one day improve people’s lives.
Aftab: Recently I published a post about evolutionary considerations around schizophrenia with a focus on a cliff-edged fitness model that had been proposed in the literature. I got some things wrong in the first post and wrote a follow-up post about clarifications and corrections. And I subsequently published a Q&A with Randolph Nesse, one of the founders of the field of evolutionary psychiatry. In the process, I was surprised by the strong skepticism some psychiatric geneticists expressed toward evolutionary hypotheses, and some reached out to me to question their scientific utility altogether. What’s your view on the relevance, or irrelevance, of evolutionary approaches to psychiatric genetics? And is there anything you'd like to add to the ongoing discussion of the evolutionary genetics of schizophrenia?
Nivard: Well, this brings us to the heart of psychiatric etiology, right? Why do some people end up in severely disturbed mental states? That “why” question might have a proximal, even individual, answer (e.g. severe trauma, a rare mutation); it might have population-level answers (sleep deprivation, precarious housing, bereavement, on average, will push some people from a state where they can cope with daily life into a state where they cannot).
When the proximal causes are genetic, it’s reasonable to ask whether an evolutionary process has, and will, change the prevalence of those genetic variants in the population. You must be careful, though, because evolutionary explanations are almost always based on assumptions about current and past environments, and about very patchy ancient DNA, if they’re based on empirical DNA at all!
But keeping that in mind, there is value in evolutionary analysis. If schizophrenia persists because of an asymmetrical fitness function (“cliff-edge fitness,” as I understand it), that can be important, as societal changes might mean more people move nearer to the cliff. If, on the other hand, there is just straightforward negative selection against any risk variant that spontaneously arises through mutation (a mutation/selection balance), that has its own set of consequences, particularly, it has consequences for how we try to find genes that are very closely related to schizophrenia. It would necessitate us to focus more on rare variant analysis.
The way I read the literature (but I am admittedly more of a bystander in this subfield) is that there is very strong evidence for negative selection. People who suffer from schizophrenia historically have fewer kids. The genetic variants that raise the risk do not become common and, in fact, disappear, while new variants arise due to mutations. Obviously, selection can act differently on individual variants or genes. The examples I have of protective copy number variants are interesting. Those variants appear to reduce risk for schizophrenia, but that risk is low to begin with, and these CNVs also have other effects that might be negative.
I go back and forth on whether we’re currently meaningfully able to model and reason about natural selection beyond the strong mutation/selection balance we observe for traits like schizophrenia. Genetic variants with strong effects on one outcome almost always affect multiple processes, making reasoning about the evolutionary consequences of a single outcome in isolation very hard.
What I’d love to find out is whether there truly is some kind of cliff edge for schizophrenia. While people with schizophrenia have far fewer children, their siblings and parents do not, but their kids do. Parents and siblings of people with the diagnosis will carry higher-than-average genetic risk but aren’t affected in their fertility. It’s truly when people end up enduring the psychosis that comes with a schizophrenia diagnosis that their fertility is reduced.
Aftab: What progress has been made in understanding gene-environment interactions? 20th century behavioral genetics with its emphasis on heritability seemed more interested in partitioning the influence of gene and environment before understanding their interaction. Has our theoretical understanding of gene-environment relationship evolved in the 21st century beyond the heritability framework?
Nivard: In many ways, we are primed to finally systematically study GxE and gene–environment correlations at scale. The most famous instance of gene–environment interaction in psychiatric genetics is potentially the G × Childhood-Trauma interaction in etiology, which, after years of hopelessly underpowered candidate gene studies and rigorous team science, turned out to be real.
Methodological innovation and scale now allow for new designs, where we don’t have to identify the specific environment we think a gene interacts with before we study GxE. This is great because, just like we thought we knew enough to do genetics research based on candidate genes but didn’t, we should probably be very modest about our ability to pick and measure the exact environments that interact with genes to contribute to the etiology of psychiatric diagnoses.
One example of a technique that doesn’t require us to predefine the E in GxE is the monozygotic twin difference test. In a recent paper, colleagues scanned the genome of MZ twins for variants where one allele led to bigger MZ differences than the other. If a gene interacts with the environment, then monozygotic twin pairs who carry that gene variant but differ in their environments across life are expected to differ more, on average, than non-carriers. We can rule out alternative explanations like G×G interaction, because MZ twins do not genetically differ. Techniques like these, and others based on similar logic, are about to allow us to study GxE at scale without pre-specifying the environment. The only limitation is that all these techniques require more from the psychiatric measures/outcomes. We must make more assumptions that require more carefully measured data, and in current massive cohort studies, there is very little appetite, among either researchers or participants, for long structured questionnaires with great psychometric properties.
I have no easy solution for the measurement problem. Researchers everywhere are struggling with dropping response rates, and the appetite for long interviews, which compete with other measurements that are equally relevant for health, just isn’t there. We’ll have to get creative. People like Rosa Cheesman and her co-authors have shown what can be accomplished when integrating genetic epidemiology with sociology and rich national register data on schools and students. She used those integrated data to identify how the genetic risk for ADHD impacts school performance, and how better schools reduce that impact, a great illustration of GxE interaction analysis, where they squeezed the most out of integrated data, and the results may have clear policy implications.
Aftab: Thank you, Michel!
This post is part of a series featuring interviews and discussions intended to foster a re-examination of philosophical and scientific debates in the psy-sciences. See prior interviews here.
See also:






From the interview, “heritability seems to matter greatly in non-scientific or popular scientific discourse” and “…most of these people really didn’t do science so much as advocacy for their views, dressed up with numbers.” This is my experience. If you read books by journalists you may be left with the impression that psychiatry is quackery; talk to sociologists and you may get the impression that psychiatric illnesses are merely social constructs; speak with social workers and you may find a strong bias toward the popular trauma narrative as an explanation for psychosis. I could go on with the biases of psychologists and criminologists as well. I think genetic determinism, mostly in scientists who haven’t stayed current in their field was already covered here. One thing all of these demonstrate is a need for more humility. This is also needed by those on the frontlines who provide social services, healthcare, and justice to those living with these illnesses and caregivers like myself who are too often confronted with ignorant and hurtful viewpoints.
Good grief the intellectual content ratio of these is huge. I wish I had something useful to say about the content, but I'm struggling to process a lot of it. Thanks for the opportunity to struggle!